Mathematical Modeling of cAMP-PDE Signaling in Atrial Fibrillation Risk Variants
Computational Biology and Complex Systems Lab, Polytechnic University of Catalonia (UPC) — Barcelona, Spain
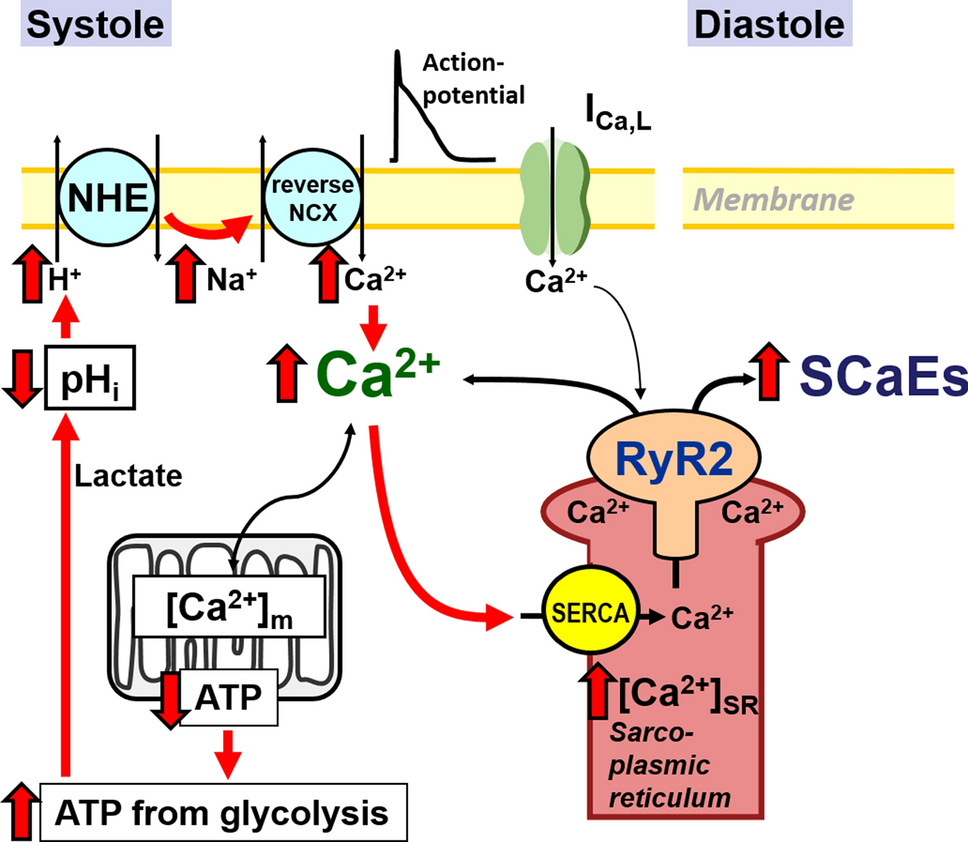
After cold emailing over 40 labs internationally, I connected with the Computational Biology and Complex Systems Lab in Barcelona. After securing a $10,000 Northwestern grant, I used this as an introduction to computational research and get a more global perspective of research. My work focused on atrial fibrillation (AF), one of the most common cardiac arrhythmias, and how disruptions in intracellular signaling could increase disease risk. I helped model the cAMP-PDE signaling pathway in atrial myocytes using MATLAB, with an emphasis on understanding the genetic risk variant rs13143308T that’s been linked to abnormal calcium levels in the heart.
I designed differential equations that represented how molecules like adenosine receptors, cyclic AMP, and phosphodiesterases interact to control calcium concentrations in heart cells. These calcium levels are a major factor in arrhythmia development. By running different simulations and testing how changes in protein concentrations affected the system, I was able to predict how this AF risk variant alters cellular behavior, even in subtle, non-obvious ways. From this project, I was able to see how computational models can help uncover the “hidden layers” of disease, which later inspired my future work.